Note
Click here to download the full example code
Visualizing decision boundaries on the P2 problem¶
This example shows the power of dynamic selection (DS) techniques which can solve complex non-linear classification near classifiers. It also compares the performance of DS techniques with some baseline classification methods such as Random Forests, AdaBoost and SVMs.
The P2 is a two-class problem, presented by Valentini, in which each class is defined in multiple decision regions delimited by polynomial and trigonometric functions:
\[\begin{split}\begin{eqnarray} \label{eq:problem1} E1(x) = sin(x) + 5 \\ \label{eq:problem2} E2(x) = (x - 2)^{2} + 1 \\ \label{eq:problem3} E3(x) = -0.1 \cdot x^{2} + 0.6sin(4x) + 8 \\ \label{eq:problem4} E4(x) = \frac{(x - 10)^{2}}{2} + 7.902 \end{eqnarray}\end{split}\]
It is impossible to solve this problem using a single linear classifier. The performance of the best possible linear classifier is around 50%.
Let’s start by importing all required modules, and defining helper functions to facilitate plotting the decision boundaries:
import matplotlib.pyplot as plt
import numpy as np
from sklearn.ensemble import AdaBoostClassifier, RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.neural_network import MLPClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
# Importing DS techniques
from deslib.dcs.ola import OLA
from deslib.dcs.rank import Rank
from deslib.des.des_p import DESP
from deslib.des.knora_e import KNORAE
from deslib.static import StackedClassifier
from deslib.util.datasets import make_P2
# Plotting-related functions
def make_grid(x, y, h=.02):
x_min, x_max = x.min() - 1, x.max() + 1
y_min, y_max = y.min() - 1, y.max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
return xx, yy
def plot_classifier_decision(ax, clf, X, mode='line', **params):
xx, yy = make_grid(X[:, 0], X[:, 1])
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
if mode == 'line':
ax.contour(xx, yy, Z, **params)
else:
ax.contourf(xx, yy, Z, **params)
ax.set_xlim((np.min(X[:, 0]), np.max(X[:, 0])))
ax.set_ylim((np.min(X[:, 1]), np.max(X[:, 0])))
def plot_dataset(X, y, ax=None, title=None, **params):
if ax is None:
ax = plt.gca()
ax.scatter(X[:, 0], X[:, 1], marker='o', c=y, s=25,
edgecolor='k', **params)
ax.set_xlabel('Feature 1')
ax.set_ylabel('Feature 2')
if title is not None:
ax.set_title(title)
return ax
Visualizing the dataset¶
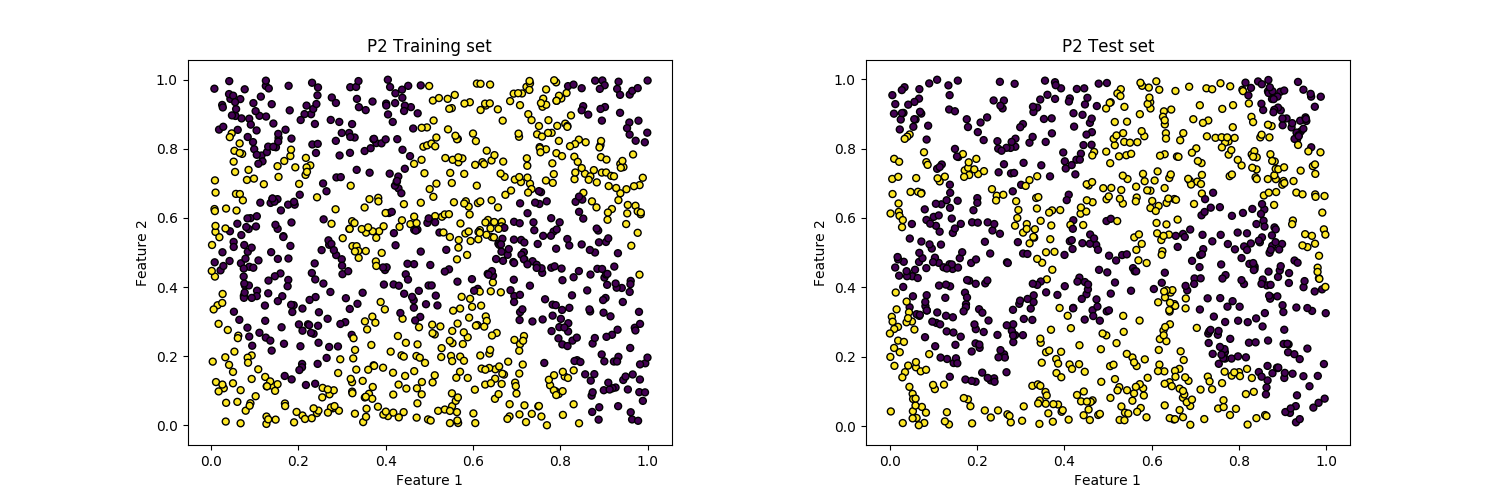
Now let’s generate and plot the dataset:
# Generating and plotting the P2 Dataset:
rng = np.random.RandomState(1234)
X, y = make_P2([1000, 1000], random_state=rng)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5,
random_state=rng)
fig, axs = plt.subplots(1, 2, figsize=(15, 5))
plt.subplots_adjust(wspace=0.4, hspace=0.4)
plot_dataset(X_train, y_train, ax=axs[0], title='P2 Training set')
plot_dataset(X_test, y_test, ax=axs[1], title='P2 Test set')
Evaluating the performance of dynamic selection methods¶
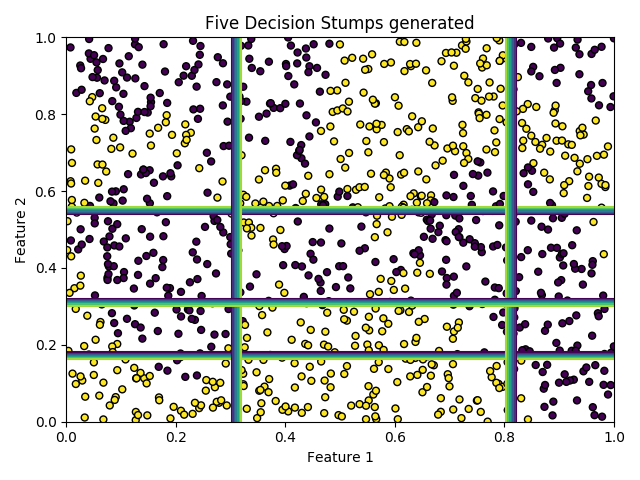
We will now generate a pool composed of 5 Decision Stumps using AdaBoost.
These are weak linear models. Each base classifier has a classification performance close to 50%.
pool_classifiers = AdaBoostClassifier(DecisionTreeClassifier(max_depth=1),
n_estimators=5, random_state=rng)
pool_classifiers.fit(X_train, y_train)
ax = plot_dataset(X_train, y_train, title='Five Decision Stumps generated')
for clf in pool_classifiers:
plot_classifier_decision(ax, clf, X_train)
ax.set_xlim((0, 1))
ax.set_ylim((0, 1))
plt.show()
plt.tight_layout()
Comparison with Dynamic Selection techniques¶
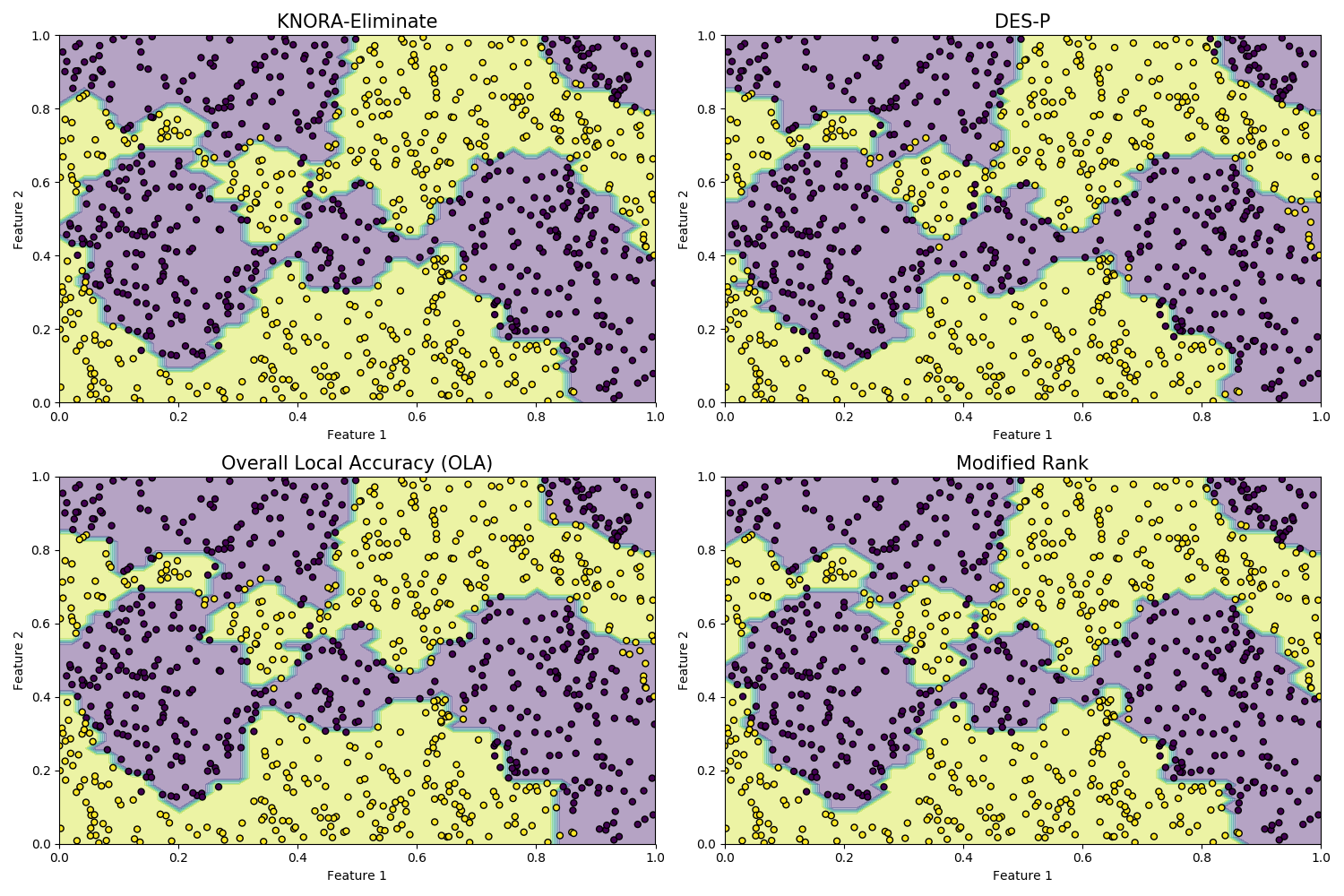
We will now consider four DS methods: k-Nearest Oracle-Eliminate (KNORA-E), Dynamic Ensemble Selection performance (DES-P), Overall Local Accuracy (OLA) and Rank. Let’s train the classifiers and plot their decision boundaries:
knora_e = KNORAE(pool_classifiers).fit(X_train, y_train)
desp = DESP(pool_classifiers).fit(X_train, y_train)
ola = OLA(pool_classifiers).fit(X_train, y_train)
rank = Rank(pool_classifiers).fit(X_train, y_train)
# Plotting the Decision Border of the DS methods.
fig2, sub = plt.subplots(2, 2, figsize=(15, 10))
plt.subplots_adjust(wspace=0.4, hspace=0.4)
titles = ['KNORA-Eliminate', 'DES-P', 'Overall Local Accuracy (OLA)',
'Modified Rank']
classifiers = [knora_e, desp, ola, rank]
for clf, ax, title in zip(classifiers, sub.flatten(), titles):
plot_classifier_decision(ax, clf, X_train, mode='filled', alpha=0.4)
plot_dataset(X_test, y_test, ax=ax)
ax.set_xlim(np.min(X[:, 0]), np.max(X[:, 0]))
ax.set_ylim(np.min(X[:, 1]), np.max(X[:, 1]))
ax.set_title(title, fontsize=15)
# Setting figure to show
# sphinx_gallery_thumbnail_number = 3
plt.show()
plt.tight_layout()
Comparison to baselines¶
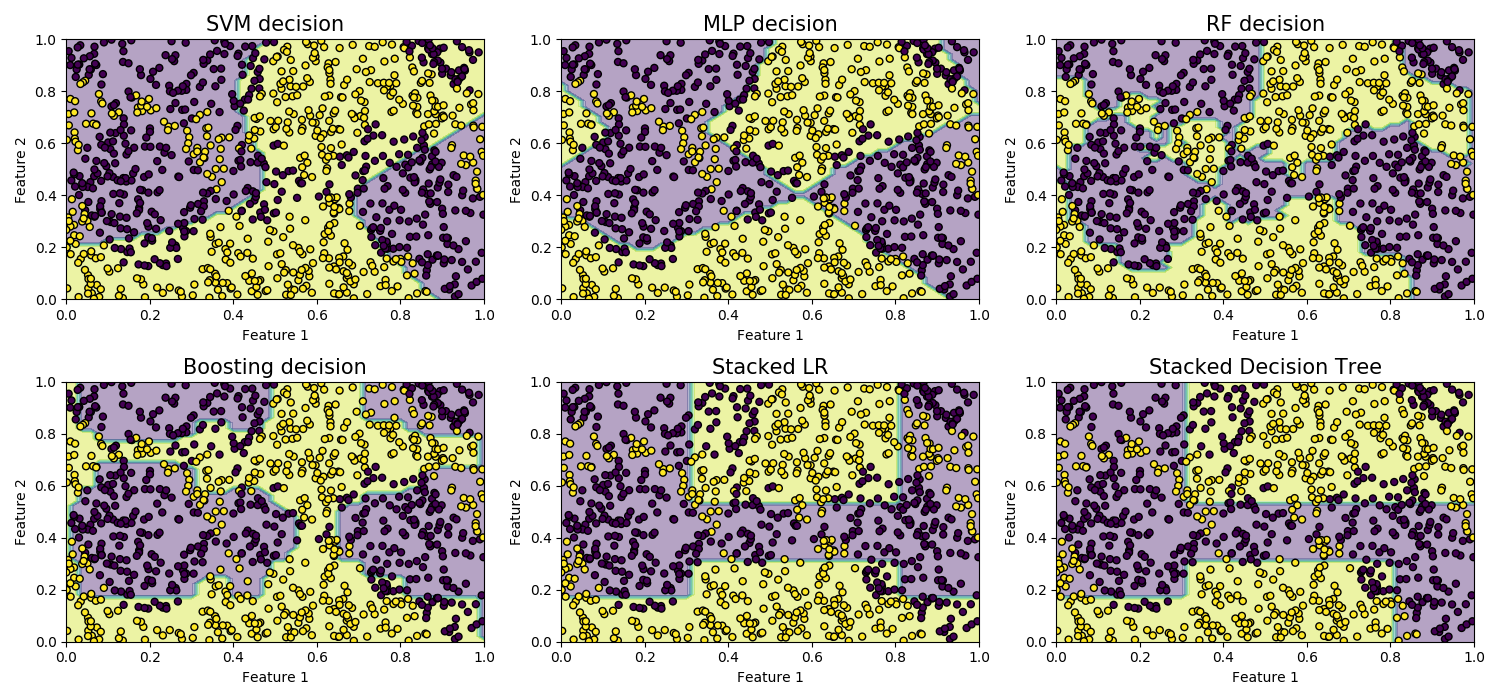
Let’s now compare the results with four baselines: Support Vector Machine (SVM) with an RBF kernel; Multi-Layer Perceptron (MLP), Random Forest, Adaboost, and Stacking.
# Setting a baseline using standard classification methods
svm = SVC(gamma='scale', random_state=rng).fit(X_train, y_train)
mlp = MLPClassifier(max_iter=10000, random_state=rng).fit(X_train, y_train)
forest = RandomForestClassifier(n_estimators=10,
random_state=rng).fit(X_train, y_train)
boosting = AdaBoostClassifier(random_state=rng).fit(X_train, y_train)
stacked_lr = StackedClassifier(pool_classifiers=pool_classifiers,
random_state=rng)
stacked_lr.fit(X_train, y_train)
stacked_dt = StackedClassifier(pool_classifiers=pool_classifiers,
random_state=rng,
meta_classifier=DecisionTreeClassifier())
stacked_dt.fit(X_train, y_train)
fig2, sub = plt.subplots(2, 3, figsize=(15, 7))
plt.subplots_adjust(wspace=0.4, hspace=0.4)
titles = ['SVM decision', 'MLP decision', 'RF decision',
'Boosting decision', 'Stacked LR', 'Stacked Decision Tree']
classifiers = [svm, mlp, forest, boosting, stacked_lr, stacked_dt]
for clf, ax, title in zip(classifiers, sub.flatten(), titles):
plot_classifier_decision(ax, clf, X_test, mode='filled', alpha=0.4)
plot_dataset(X_test, y_test, ax=ax)
ax.set_xlim(np.min(X[:, 0]), np.max(X[:, 0]))
ax.set_ylim(np.min(X[:, 1]), np.max(X[:, 1]))
ax.set_title(title, fontsize=15)
plt.show()
plt.tight_layout()
Evaluation on the test set¶
Finally, let’s evaluate the baselines and the Dynamic Selection methods on the test set:
print('KNORAE score = {}'.format(knora_e.score(X_test, y_test)))
print('DESP score = {}'.format(desp.score(X_test, y_test)))
print('OLA score = {}'.format(ola.score(X_test, y_test)))
print('Rank score = {}'.format(rank.score(X_test, y_test)))
print('SVM score = {}'.format(svm.score(X_test, y_test)))
print('MLP score = {}'.format(mlp.score(X_test, y_test)))
print('RF score = {}'.format(forest.score(X_test, y_test)))
print('Boosting score = {}'.format(boosting.score(X_test, y_test)))
print('Stacking LR score = {}' .format(stacked_lr.score(X_test, y_test)))
print('Staking Decision Tree = {}' .format(stacked_dt.score(X_test, y_test)))
Out:
KNORAE score = 0.948
DESP score = 0.927
OLA score = 0.932
Rank score = 0.948
SVM score = 0.725
MLP score = 0.794
RF score = 0.923
Boosting score = 0.795
Stacking LR score = 0.716
Staking Decision Tree = 0.732
Total running time of the script: ( 0 minutes 18.277 seconds)