Note
Click here to download the full example code
Comparing dynamic selection with Random Forest¶
In this example we use a pool of classifiers generated using the Random Forest method rather than Bagging. We also show how to change the size of the region of competence, used to estimate the local competence of the base classifiers.
This demonstrates that the library accepts any kind of base classifiers as long as they implement the predict and predict proba functions. Moreover, any ensemble generation method such as Boosting or Rotation Trees can be used to generate a pool containing diverse base classifiers. We also included the performance of the RandomForest classifier as a baseline comparison.
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.cm import get_cmap
from matplotlib.ticker import FuncFormatter
from sklearn.datasets import fetch_openml
# Pool of base classifiers
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from deslib.dcs.mcb import MCB
# Example of a dcs techniques
from deslib.dcs.ola import OLA
# Example of a des techniques
from deslib.des.des_p import DESP
from deslib.des.knora_e import KNORAE
from deslib.des.knora_u import KNORAU
from deslib.des.meta_des import METADES
# Example of stacked model
from deslib.static.stacked import StackedClassifier
rng = np.random.RandomState(42)
# Fetch a classification dataset from OpenML
data = fetch_openml(name='credit-g', version=1, cache=False, as_frame=False)
X = data.data
y = data.target
# split the data into training and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25,
random_state=rng)
# Training a random forest to be used as the pool of classifiers.
# We set the maximum depth of the tree so that it
# can estimate probabilities
RF = RandomForestClassifier(random_state=rng, n_estimators=10)
RF.fit(X_train, y_train)
X_train, X_dsel, y_train, y_dsel = train_test_split(X_train, y_train,
test_size=0.50,
random_state=rng)
stacked = StackedClassifier(RF, LogisticRegression())
stacked.fit(X_dsel, y_dsel)
# Initialize a DS technique. Here we specify the size of
# the region of competence (5 neighbors)
knorau = KNORAU(RF, k=5, random_state=rng)
kne = KNORAE(RF, k=5, random_state=rng)
desp = DESP(RF, k=5, random_state=rng)
ola = OLA(RF, k=5, random_state=rng)
mcb = MCB(RF, k=5, random_state=rng)
meta = METADES(RF, k=5, random_state=rng)
# Fit the DS techniques
knorau.fit(X_dsel, y_dsel)
kne.fit(X_dsel, y_dsel)
desp.fit(X_dsel, y_dsel)
meta.fit(X_dsel, y_dsel)
ola.fit(X_dsel, y_dsel)
mcb.fit(X_dsel, y_dsel)
Out:
MCB(k=5,
pool_classifiers=RandomForestClassifier(n_estimators=10,
random_state=RandomState(MT19937) at 0x7F883E86F888),
random_state=RandomState(MT19937) at 0x7F883E86F888)
Plotting the results¶
Let’s now evaluate the methods on the test set.
rf_score = RF.score(X_test, y_test)
stacked_score = stacked.score(X_test, y_test)
knorau_score = knorau.score(X_test, y_test)
kne_score = kne.score(X_test, y_test)
desp_score = desp.score(X_test, y_test)
ola_score = ola.score(X_test, y_test)
mcb_score = mcb.score(X_test, y_test)
meta_score = meta.score(X_test, y_test)
print('Classification accuracy RF: ', rf_score)
print('Classification accuracy Stacked: ', stacked_score)
print('Evaluating DS techniques:')
print('Classification accuracy KNORA-U: ', knorau_score)
print('Classification accuracy KNORA-E: ', kne_score)
print('Classification accuracy DESP: ', desp_score)
print('Classification accuracy OLA: ', ola_score)
print('Classification accuracy MCB: ', mcb_score)
print('Classification accuracy META-DES: ', meta_score)
cmap = get_cmap('Dark2')
colors = [cmap(i) for i in np.linspace(0, 1, 7)]
labels = ['RF', 'Stacked', 'KNORA-U', 'KNORA-E', 'DESP', 'OLA', 'MCB',
'META-DES']
fig, ax = plt.subplots()
pct_formatter = FuncFormatter(lambda x, pos: '{:.1f}'.format(x * 100))
ax.bar(np.arange(8),
[rf_score, stacked_score, knorau_score, kne_score, desp_score,
ola_score, mcb_score, meta_score],
color=colors,
tick_label=labels)
ax.set_ylim(0.65, 0.80)
ax.set_xlabel('Method', fontsize=13)
ax.set_ylabel('Accuracy on the test set (%)', fontsize=13)
ax.yaxis.set_major_formatter(pct_formatter)
for tick in ax.get_xticklabels():
tick.set_rotation(45)
plt.subplots_adjust(bottom=0.15)
plt.show()
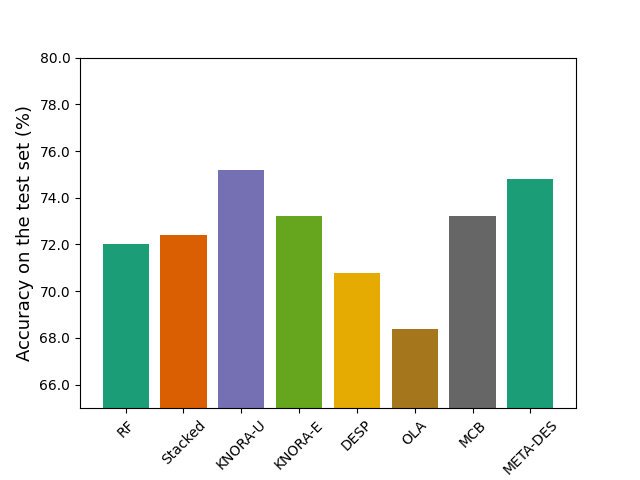
Out:
Classification accuracy RF: 0.72
Classification accuracy Stacked: 0.724
Evaluating DS techniques:
Classification accuracy KNORA-U: 0.752
Classification accuracy KNORA-E: 0.732
Classification accuracy DESP: 0.708
Classification accuracy OLA: 0.684
Classification accuracy MCB: 0.732
Classification accuracy META-DES: 0.748
Total running time of the script: ( 0 minutes 6.074 seconds)