Note
Click here to download the full example code
Dynamic selection vs K-NN: Using instance hardness¶
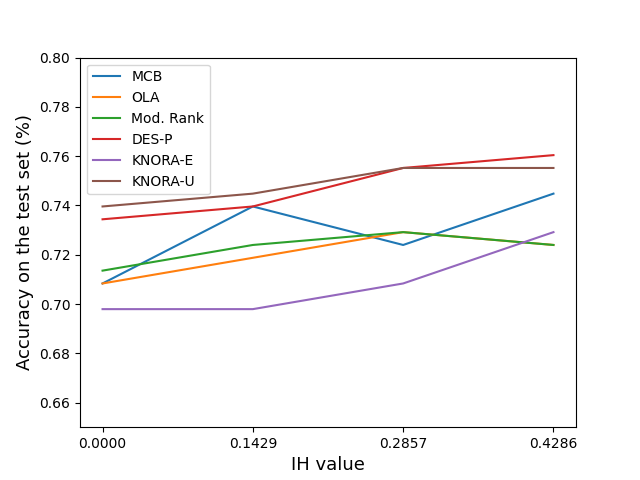
One aspect about dynamic selection techniques is that it can better deal with the classification of test examples associated with high degree of instance hardness. Such examples are often found close to the border of the classes, with the majority of its neighbors belonging to different classes. On the other hand, the KNN method, which is often used to estimate the region of competence in DS methods works better in the classification of examples associated with low instance hardness [1].
DESlib already implements a switch mechanism between DS techniques and the KNN classifier according to the hardness level of an instance. This example varies the threshold in which KNN is used for classification instead of DS methods. It also compares the classification results with the standard KNN as a baseline.
The switch mechanism also reduces the computational cost involved since only part of the test samples are classified by the DS method.
References¶
[1] Cruz, Rafael MO, et al. “Dynamic Ensemble Selection VS K-NN: why and when Dynamic Selection obtains higher classification performance?.” arXiv preprint arXiv:1804.07882 (2018).
Let’s start by importing all required modules. In this example we use the new sklearn-OpenML interface to fetch the diabetes classification problem.
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import fetch_openml
from sklearn.ensemble import BaggingClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.tree import DecisionTreeClassifier
from deslib.dcs import MCB
from deslib.dcs import OLA
from deslib.dcs import Rank
from deslib.des import DESP
from deslib.des import KNORAE
from deslib.des import KNORAU
rng = np.random.RandomState(123456)
data = fetch_openml(name='diabetes', version=1, cache=False, as_frame=False)
X = data.data
y = data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=rng)
# Normalizing the dataset to have 0 mean and unit variance.
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
# Training a pool of classifiers using the bagging technique.
pool_classifiers = BaggingClassifier(DecisionTreeClassifier(random_state=rng),
random_state=rng)
pool_classifiers.fit(X_train, y_train)
Out:
BaggingClassifier(base_estimator=DecisionTreeClassifier(random_state=RandomState(MT19937) at 0x7F883E86F570),
random_state=RandomState(MT19937) at 0x7F883E86F570)
Setting DS method to use the switch mechanism¶
In order to activate the functionality to switch between DS and KNN according to the instance hardness level we need to set the DS techniques to use this information. This is done by setting the hyperparameter with_IH to True. In this example we consider four different values for te threshold
mcb = MCB(pool_classifiers, with_IH=True, random_state=rng)
ola = OLA(pool_classifiers, with_IH=True, random_state=rng)
rank = Rank(pool_classifiers, with_IH=True, random_state=rng)
des_p = DESP(pool_classifiers, with_IH=True, random_state=rng)
kne = KNORAE(pool_classifiers, with_IH=True, random_state=rng)
knu = KNORAU(pool_classifiers, with_IH=True, random_state=rng)
list_ih_values = [0.0, 1./7., 2./7., 3./7.]
list_ds_methods = [method.fit(X_train, y_train) for method in
[mcb, ola, rank, des_p, kne, knu]]
names = ['MCB', 'OLA', 'Mod. Rank', 'DES-P', 'KNORA-E', 'KNORA-U']
# Plot accuracy x IH
fig, ax = plt.subplots()
for ds_method, name in zip(list_ds_methods, names):
accuracy = []
for idx_ih, ih_rate in enumerate([0.0, 0.14, 0.28, 0.42]):
ds_method.IH_rate = ih_rate
accuracy.append(ds_method.score(X_test, y_test))
ax.plot(list_ih_values, accuracy, label=name)
plt.xticks(list_ih_values)
ax.set_ylim(0.65, 0.80)
ax.set_xlabel('IH value', fontsize=13)
ax.set_ylabel('Accuracy on the test set (%)', fontsize=13)
ax.legend()
plt.show()
Total running time of the script: ( 0 minutes 7.586 seconds)